
Hi! I’m Laura Toribio (a.k.a. Tori). I’m a Bioinformatician with a nearly completed PhD and over 4 years of hands-on experience in analyzing multi-omics biological data. I’m competent in Python, R and shell scripting, continuously enhancing my programming skills. Also hooked on aesthetic data visualization and presentation.
My growing interest for emerging technologies has led me to explore and learn Data Science. My aspiration is to merge Bioinformatics and Machine Learning, applied to biological processes, to foster progress in healthcare and life sciences.
Technical skills
- Python: pandas, numpy, matplotlib, plotly
- Data Science: scikit-learn, TensorFlow, Keras
- Shell scripting: bash, awk, sed
- Containerization: Docker
- Amazon Web Services: AWS Lambda, API Gateway, Elastic Container Registry
- High Performance Computing (HPC)
- R: ggplot2, tidyverse
- Git & Github
Projects
-
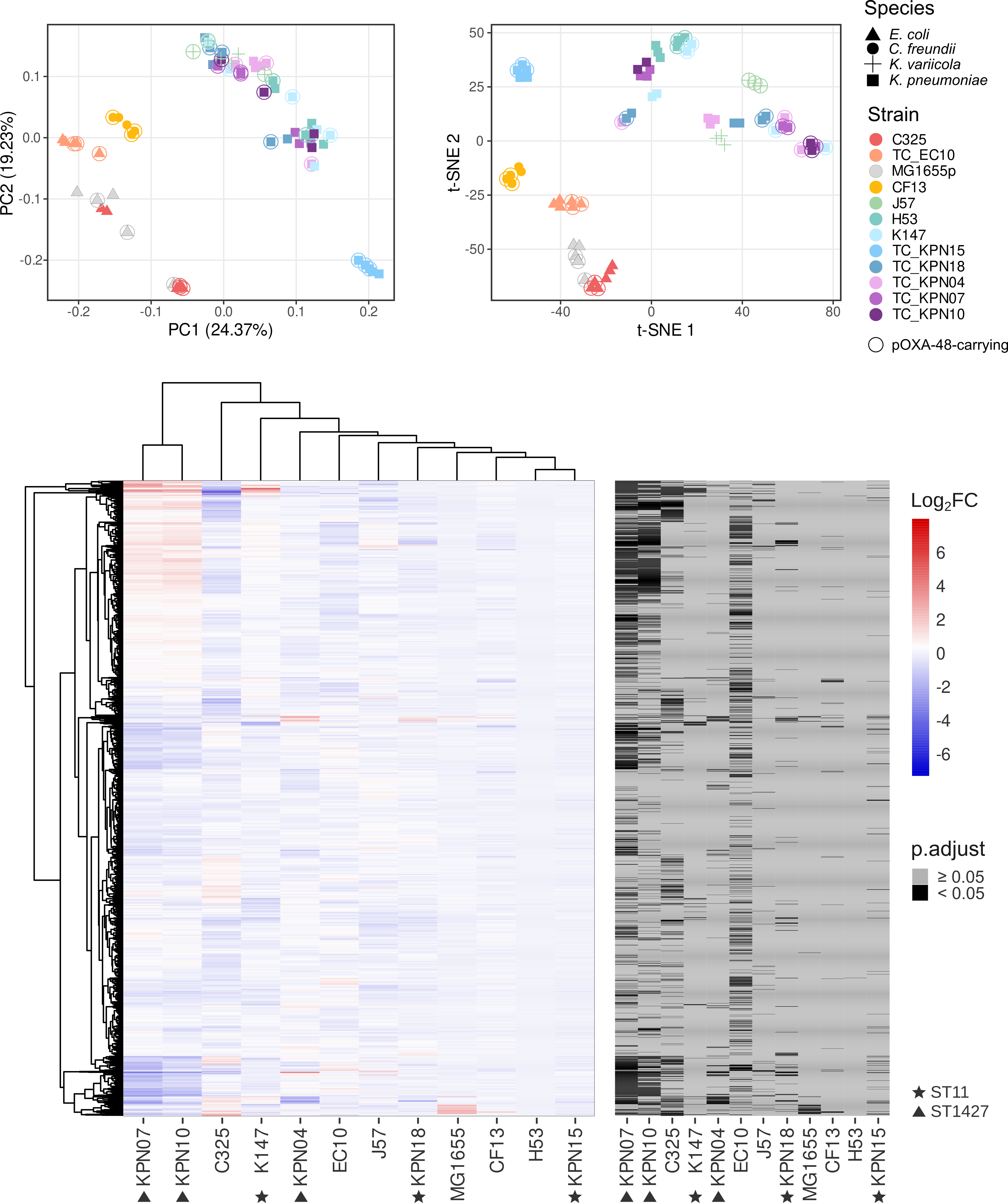
Dimensionality Reduction and Hierarchical Clustering
- Performed PCA, t-SNE and Hierarchical Clustering in R to identify clusters of bacterial strains with similar transcriptional profiles.
- Analyses form part of the publication: Toribio-Celestino et al. (2024) Nature Communications.
- Repository link: Dimensionality reduction and hierarchical clustering
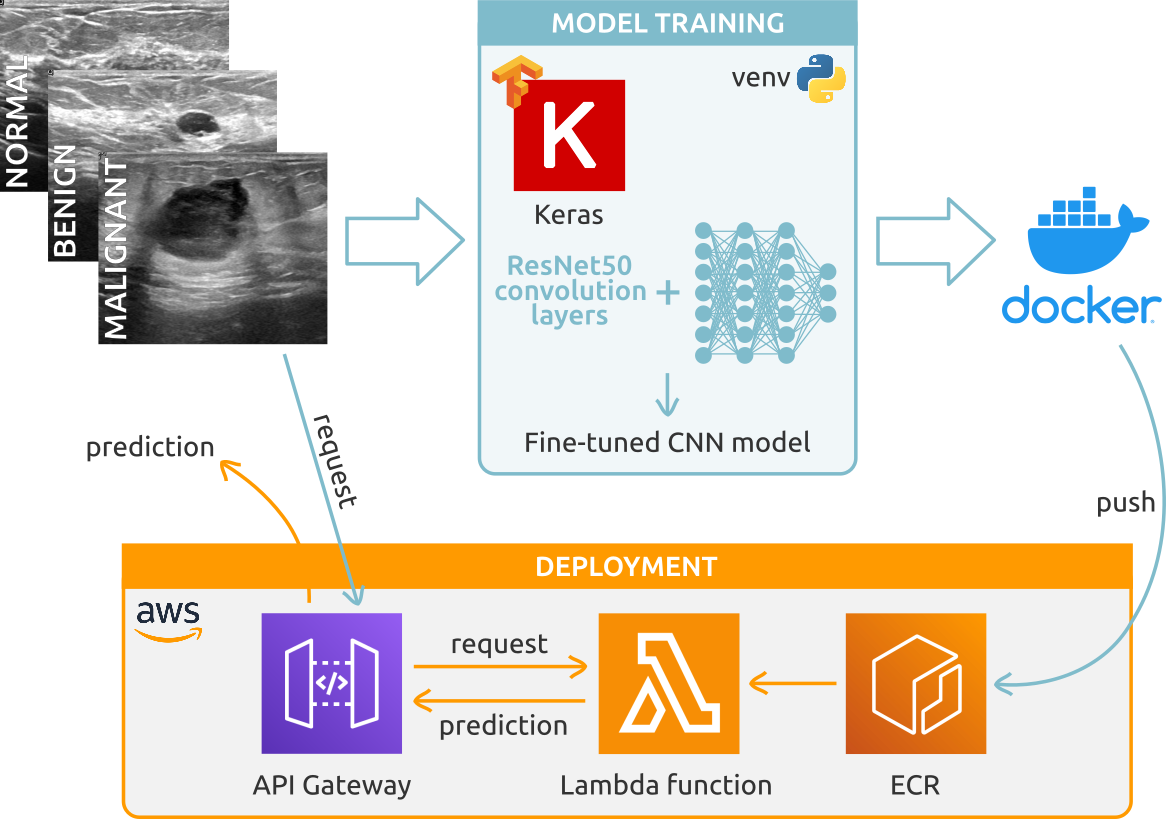
Breast Cancer Image Classifier
- Developed a Convolutional Neural Network model to classify breast ultrasound images into normal, benign and malignant, achieving an overall 85% accuracy.
- Performed transfer learning using TensorFlow and Keras, training a Neural Network on top of the ResNet50 architecture, with subsequent fine-tuning of the outer convolution layers to improve model accuracy.
- Deployed the model as a web service through AWS Lambda and API Gateway, with the containerized model (Docker) uploaded in a AWS Elastic Container Registry.
- Repository link: Breast Cancer Image Classifier
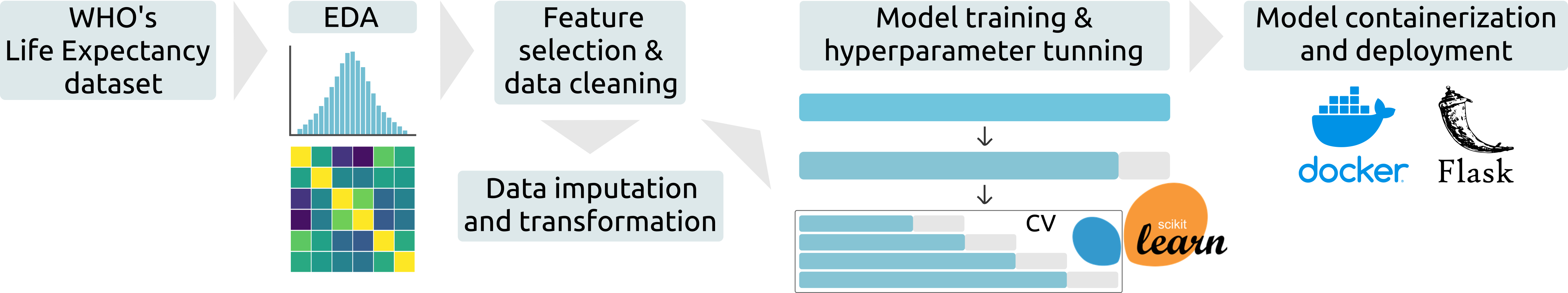
Life Expectancy Prediction
- Deployed a robust regression model to predict the life expectancy of a population, achieving a high model accuracy, with an average prediction error of 2.8 years.
- Performed exploratory data analysis and preprocessing (feature selection, missing data imputation with MICE, data transformation, etc) of the WHO’s Life Expectancy dataset, improving the dataset quality.
- Trained and simultaneously fine-tuned different ML models (linear regressions, SVM, decision trees and ensemble methods), using scikit-learn, to predict the life expectancy of a given country, accounting for the time-series nature of the dataset.
- Model containerization and deployment with Docker and Flask.
- Repository link: Life Expectancy Prediction
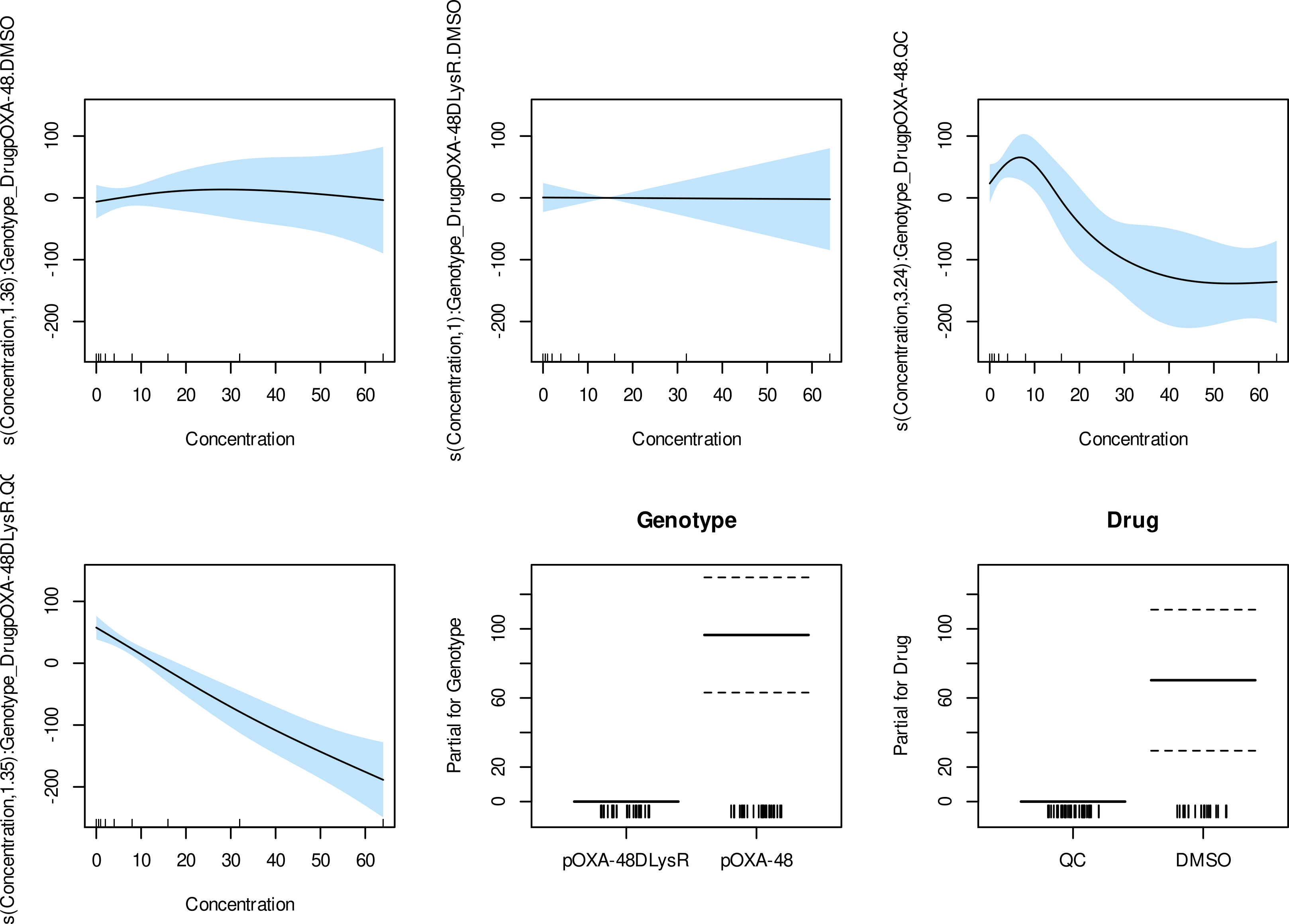
Generalized Additive Models (GAMs)
- Constructed two GAMs with the R package mgcv to assess the effect of the concentration of two treatments in the growth (measured as AUC of growth curves) of two bacterial strains, including interaction terms for the type of treatment and the genotype.
- Analyses form part of the publication: Toribio-Celestino et al. (2024) Nature Communications.
- Repository link: GAMs
-
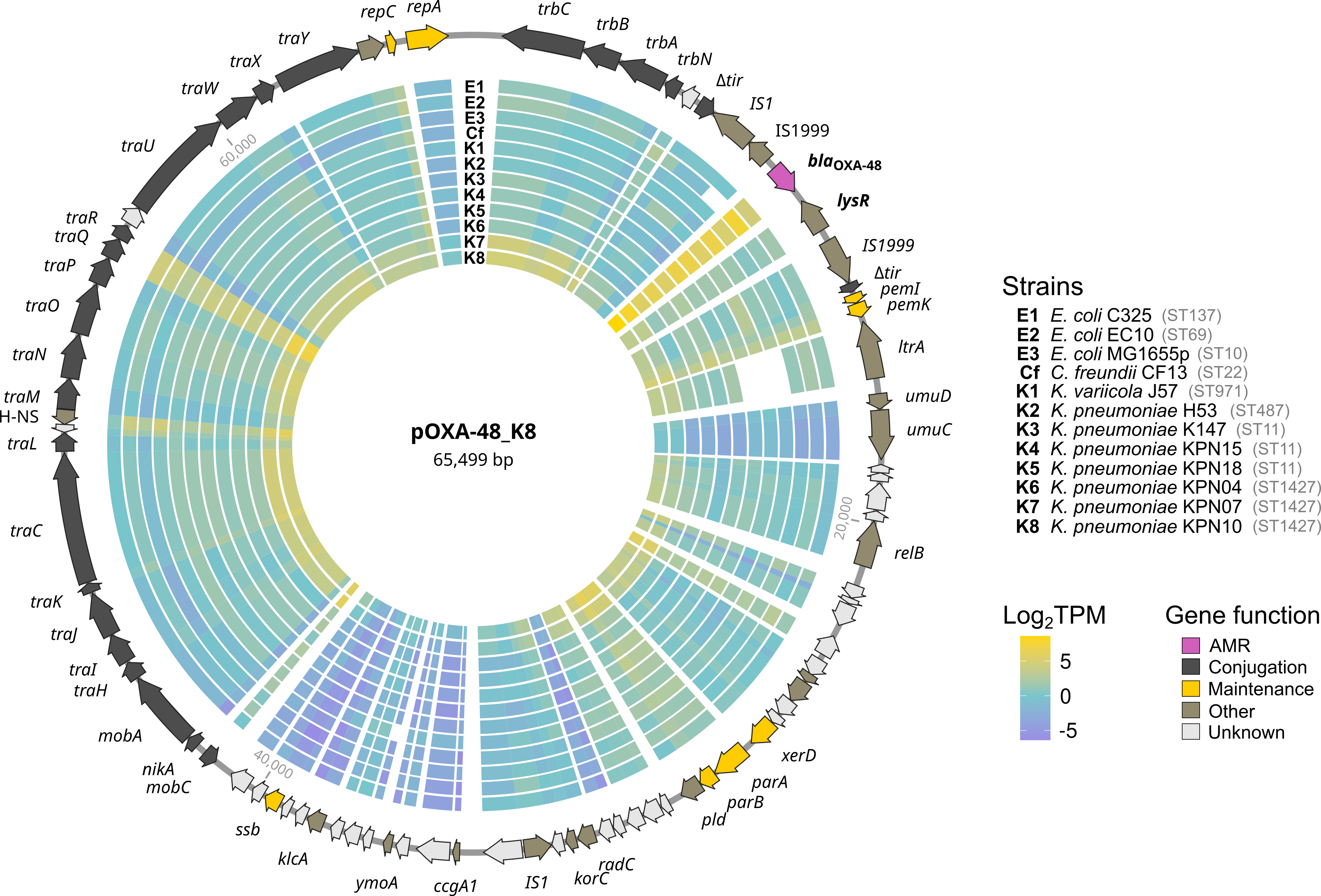
Transcriptomic Analyses
- Developed a standard workflow for RNA-Seq data analysis in R, including read count with featureCounts and differential expression analysis with DESeq2 (see Hierarchichal Clustering).
- Performed gene set enrichment analysis (GSEA) with clusterProfiler.
- Wrote Shell (documented in MarkDown files) and Python scripts for manipulating/filtering output files used as input in downstream steps of the pipeline.
- Analyses form part of the publications: Toribio-Celestino et al. (2024) Nature Communications, Blanco et al. (2024) Nucleic Acids Research, Sastre-Dominguez et al. (2024) Nature Ecology and Evolution and Blanco et al. (2024) Nucleic Acids Research.
- Repository links: RNA-Seq_1st_dataset, RNA-Seq_DeltaLysR and RNA-Seq_experimental_evolution
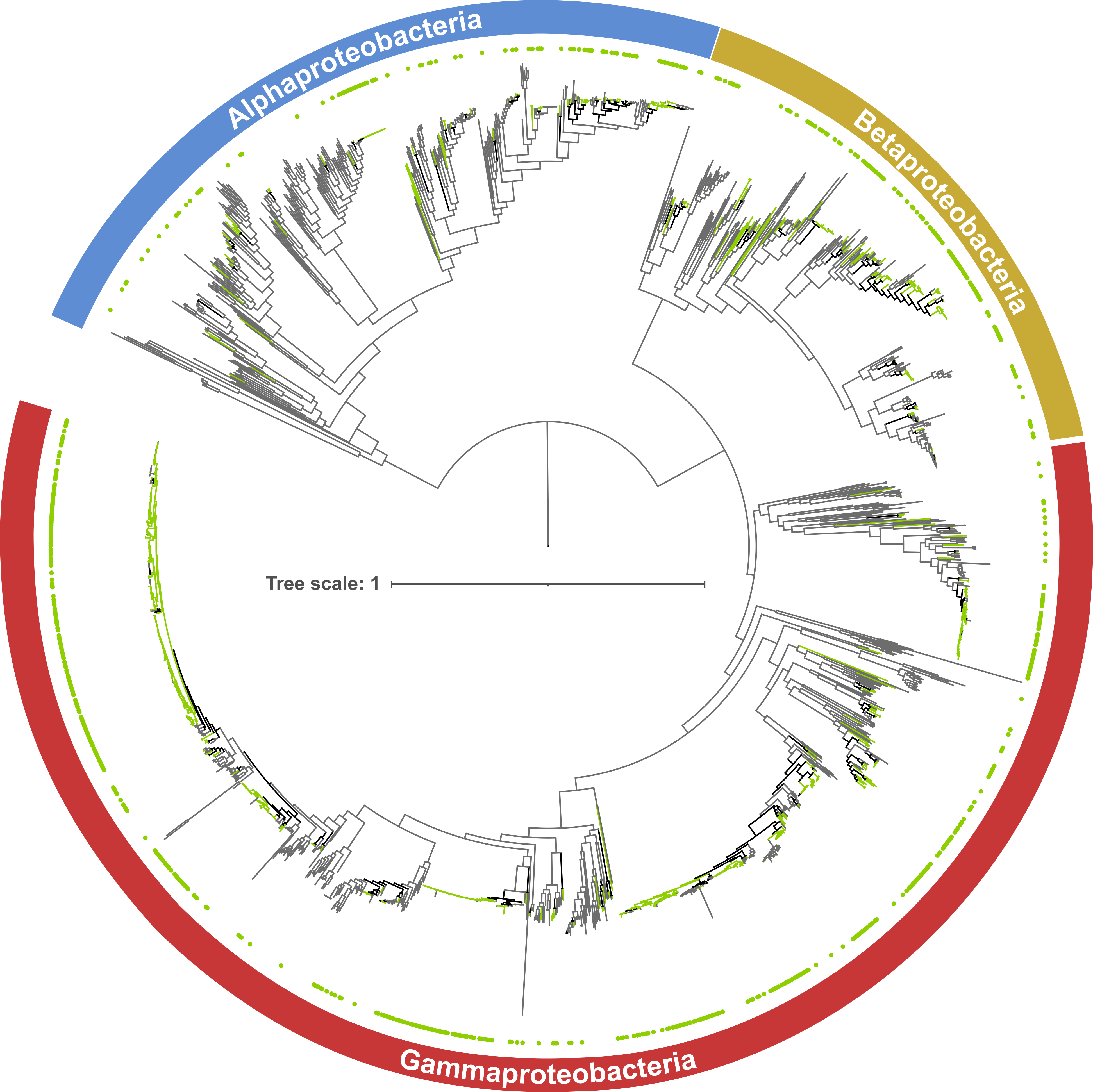
Ancestral Character Reconstruction
- Downloaded and analyzed a large RefSeq database of bacterial genomes.
- Constructed large bacterial phylogenies using IQ-TREE from genetic markers identified with Hidden Markov Models (hmmsearch).
- Performed ancestral character reconstruction with PastML, gaining insights into the ancestry of a genetic trait.
- Wrote Shell (documented in MarkDown files) and Python scripts for manipulating/filtering output files used as input in downstream steps of the pipeline.
- Analyses form part of the publication: Toribio-Celestino et al. (2024) Nature Communications.
- Repository link: LysR_and_the_mystery_operon
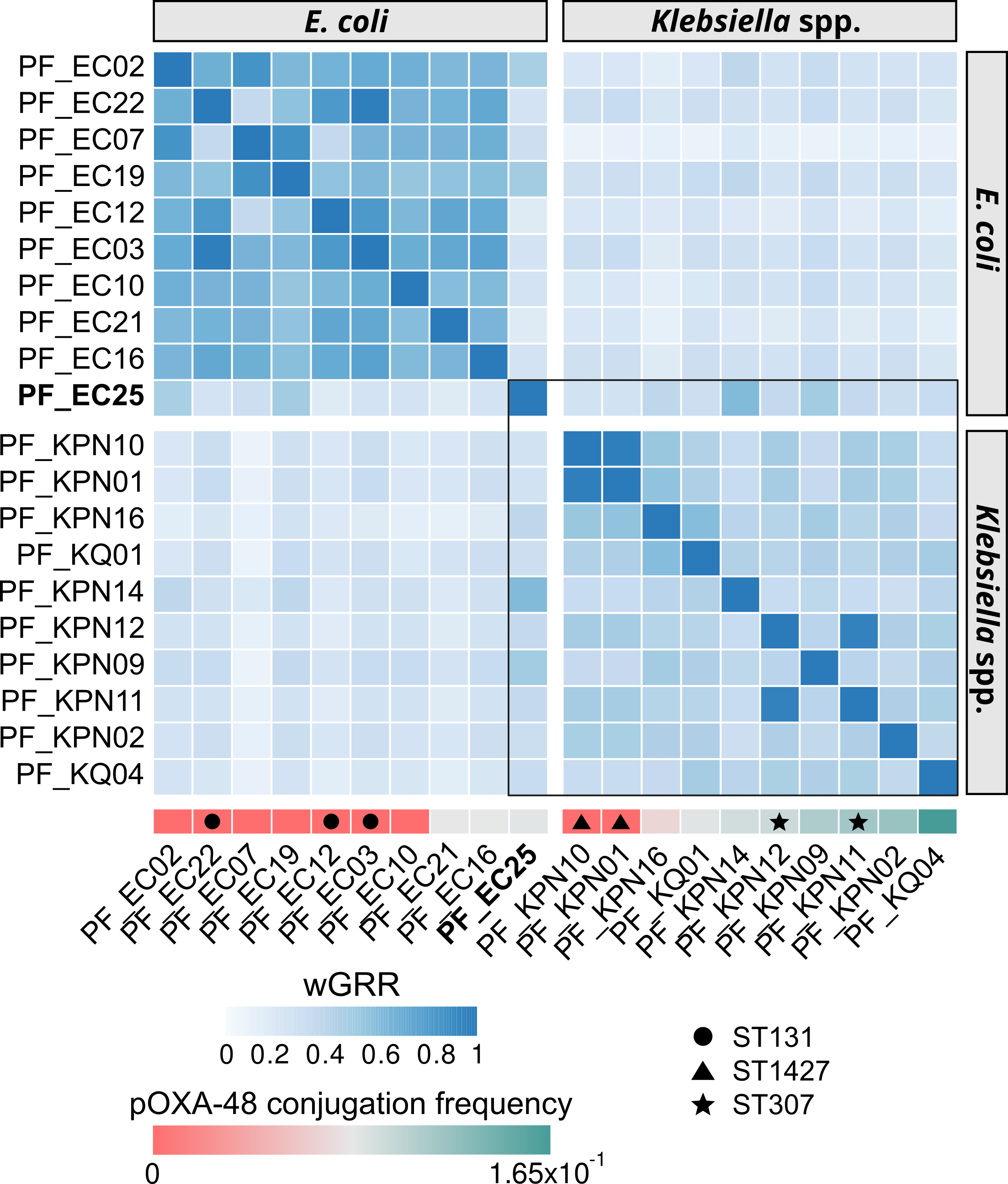
Comparative Genomic Analysis and Phylogenetic Logistic Regression
- Performed diverse comparative genomic analyses to identify genetic traits that could be associated to a certain phenotype, using tools like MacSyFinder or BLAST.
- Analyzed the potential associations with Fisher’s exact tests and phylogenetic logistic regression (phyloglm in R) to account for phylogenetic dependency.
- Wrote Shell (documented in MarkDown files) and Python scripts for manipulating/filtering output files used as input in downstream steps of the pipeline.
- Analyses form part of the publication: Alonso-del Valle et al. (2023) PNAS.
- Repository link: Super-sinks
Variant Calling
- Constructed de novo genomic assemblies using SPAdes, Unicycler and Flye.
- Performed diverse analyses of genomic variants using tools like snippy and breseq.
- Analyses form part of the publications: Toribio-Celestino et al. (2024) Nature Communications, Fernández-Calvet et al. (2023) Microbiology and DelaFuente et al. (2022) Nature Ecology and Evolution.
- Repository links: Closing_reference_genomes, Within-patient_evolution and CRISPR_cured_pOXA-48

Experience
Bioinformatics Scientist @ Centro Nacional de Biotecnología–CSIC, Madrid, Spain (Feb. 2021 – Present)
Visiting Scientist @ Institut Pasteur, Paris, France (Feb. 2022 – Mar. 2022)
Junior Bioinformatics Scientist @ IRYCIS, Madrid, Spain (Oct. 2019 – Feb. 2021)
Research Assistant @ Ikan Biotech, Noain, Spain (Nov. 2018 – Jun. 2019)
Junior Research Scientist @ Centro de Biología Molecular Severo Ochoa–CSIC, Madrid, Spain (Feb. 2018 – Sept. 2018)
Junior Clinical Scientist @ Hospital Universitario de Guadalajara, Guadalajara, Spain (Feb. 2017 – Jun. 2017)
Education
PhD in Molecular Biosciences – Bioinformatics and Systems Biology @ Universidad Autónoma de Madrid, Spain (2020 – Present)
MSc in Advanced Bioinformatic Analysis – NGS Data Analysis and Data Science @ Universidad Pablo de Olavide de Sevilla, Spain (2018 – 2021)
MSc in Biotechnology @ Universidad Autónoma de Madrid, Spain (2017 – 2018)
BSc in Biology @ Universidad de Alcalá de Henares, Spain (2012 – 2017)
Publications
- Toribio-Celestino L#, San Millan A. (2025) Plasmid–bacteria associations in the clinical context. Trends Microbiol. In press.
- Toribio-Celestino L*, Calvo-Villamañán A*, Herencias C, Alonso-Del Valle A, Sastre-Dominguez J, Quesada S, Mazel D, Rocha EPC, Fernández-Calvet A, San Millan A. (2024) A plasmid–chromosome crosstalk in multidrug resistant enterobacteria. Nat Commun. 15(1):10859. doi: 10.1038/s41467-024-55169-y.
- Blanco P, Trigo da Roza F, Toribio-Celestino L, García-Pastor L, Caselli N, Morón Á, Ojeda F, Darracq B, Vergara E, Amaro F, San Millan A, Skovgaard O, Mazel D, Loot C, Escudero JA. (2024) Chromosomal integrons are genetically and functionally isolated units of genomes. Nucleic Acids Res. 52(20):12565-12581. doi: 10.1093/nar/gkae866.
- Sastre-Dominguez J, DelaFuente J, Toribio-Celestino L, Herencias C, Herrador-Gomez P, Costas C, Hernandez-Garcia M, Canton R, Rodriguez-Beltran J, Santos-Lopez A, San Millan A. (2024) Plasmid-encoded insertion sequences promote rapid adaptation in clinical enterobacteria. Nat Ecol Evol. 8(11):2097-2112. doi: 10.1038/s41559-024-02523-4.
- Blanco P, Hipólito A*, García-Pastor L*, Trigo da Roza F, Toribio-Celestino L, Ortega AC, Vergara E, San Millán Á, Escudero JA. (2024) Identification of promoter activity in gene-less cassettes from Vibrionaceae superintegrons. Nucleic Acids Res. 52(6):2961-2976. doi: 10.1093/nar/gkad1252.
- Alonso-Del Valle A*, Toribio-Celestino L*, Quirant A, Pi CT, DelaFuente J, Canton R, Rocha EPC, Ubeda C, Peña-Miller R, San Millan A. (2023) Antimicrobial resistance level and conjugation permissiveness shape plasmid distribution in clinical enterobacteria. Proc Natl Acad Sci USA. 120(51):e2314135120. doi: 10.1073/pnas.2314135120.
- Fernández-Calvet A*, Toribio-Celestino L*, Alonso-Del Valle A, Sastre-Dominguez J, Valdes-Chiara P, San Millan A, DelaFuente J. (2023) The distribution of fitness effects of plasmid pOXA-48 in clinical enterobacteria. Microbiology. 169(7):001369. doi: 10.1099/mic.0.001369.
- DelaFuente J, Toribio-Celestino L, Santos-Lopez A, León-Sampedro R, Alonso-Del Valle A, Costas C, Hernández-García M, Cui L, Rodríguez-Beltrán J, Bikard D, Cantón R, San Millan A. (2022) Within-patient evolution of plasmid-mediated antimicrobial resistance. Nat Ecol Evol. 6(12):1980-1991. doi: 10.1038/s41559-022-01908-7.
# Corresponding author
* Contributed equally
